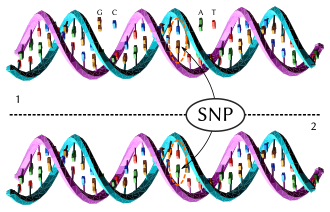
Single nucleotide polymorphism
A single nucleotide polymorphism (SNP, pronounced snip; plural snips) is a DNA sequence variation in a population. A SNP is just a single nucleotide difference in the genome.
For example, sequenced DNA fragments from two people, AAGCCTA to AAGCTTA, is different in a single nucleotide. In this case we say that there are two alleles. Almost all common SNPs have only two alleles. SNPs occur most often in regions of the DNA which do not affect the survival of the organism: otherwise they would be weeded out by natural selection.[1] Other factors, like genetic recombination and mutation rate, can also affect SNP density.[2] There are variations between human populations, so a SNP allele that is common in one geographical or ethnic group may be much rarer in another.
These genetic variations between individuals (particularly in non-coding parts of the genome) are sometimes exploited in DNA fingerprinting, which is used in forensic science. Also, these genetic variations cause differences in our susceptibility to disease. The severity of illness and the way our body responds to treatments are also manifestations of genetic variations. For example, a single base mutation in the APOE (apolipoprotein E) gene is associated with a higher risk for Alzheimer's disease.[3]
Single Nucleotide Polymorphism Media
References
- ↑ Barreiro L.B. et al 2008 (2008). "Natural selection has driven population differentiation in modern humans". Nature Genetics. 40 (3): 340–345. doi:10.1038/ng.78. PMID 18246066. S2CID 205357396.
- ↑ Nachman, Michael W. (2001). "Single nucleotide polymorphisms and recombination rate in humans". Trends in Genetics. 17 (9): 481–485. doi:10.1016/S0168-9525(01)02409-X. PMID 11525814.
- ↑ Wolf A.B. et al 2012. APOE and neuroenergetics: an emerging paradigm in Alzheimer's disease. Neurobiology of Aging 34 (4): 1007–17. [1]